J.ophthalmol.(Ukraine).2020;2:3-11.
|
http://doi.org/10.31288/oftalmolzh20202311 Received: 15 January 2020; Published on-line: 30 April 2020 Role of IL6 -174 G/C, IL10 1082G/A and IL10 -592C/A in the pathogenesis of keratoconus and development of recurrent erosion in Ukrainian patients with lattice corneal dystrophy Livshits L.A.1, Dr Sc (Med), Prof.; Drozhzhyna G.I.2, Dr Sc (Med), Prof.; Kucherenko A.M.1; Ivanovska O.V.1; Gaidamaka T.B.1, Dr Sc (Med); Gorodna O.V.1; Sereda K.V.2 1 Institute of Molecular Biology and Genetics, National Academy of Sciences of Ukraine; Kyiv (Ukraine) 2 Filatov Institute of Eye Diseases and Tissue Therapy, National Academy of Medical Science of Ukraine; Odesa (Ukraine) E-mail: livshits@edu.imbg.org.ua TO CITE THIS ARTICLE: Livshits LA, Drozhzhyna GI, Kucherenko AM, Ivanovska OV, Gaidamaka TB, Gorodna OV, Sereda K.V. Role of IL-6 -174 G/C, 1082G/A and IL-10 -592C/A in the pathogenesis of keratoconus and development of recurrent erosion in Ukrainian patients with lattice corneal dystrophy. J.ophthalmol.(Ukraine).2020;2:3-11. http://doi.org/10.31288/oftalmolzh20202311
Background: Keratoconus (KC, or corneal ectasia) is a multifactorial disease with a genetic component and an average annual incidence rate of 2.0/100,000 persons. Lattice corneal stromal dystrophy (LCD), a monogenic disorder with varying phenotypic manifestations, is the most common hereditary corneal dystrophy associated with mutations in the TGFBI gene in Ukraine, with as much as 40.2% of cases attributed to this disease. Purpose: To elucidate the role of polymorphic variants in the IL6 promoter (-174 G/C) and IL10 (-1082G/A and -592C/A) as factors of a genetic predisposition to KC and recurrent corneal erosion in Ukrainian patients with LCD. Material and Methods: All patients underwent a routine eye examination including visual acuity assessment, biomicroscopy, fluorescein testing, tonometry and ophthalmoscopy. In addition, patients with KC underwent keratotopography, pachymetry, remote biometry and gonioscopy. Genotyping was done for IL6 -174 G/C, IL10 -1082G/A and IL10 -592C/A by polymerase chain reaction followed by restriction fragment length polymorphism. Fexact test was used for statistical analyses. Results: The frequency of homozygotes (AA) for IL10 rs1800896 was increased, whereas the frequency of homozygotes (CC) for IL6 174G/С was decreased in patients with KC compared to controls (0.25 vs 0.19 and 0.18 vs 0.22, respectively), although the differences were not statistically significant. The frequency of IL6 C allele carriers was significantly higher among patients with LCD and recurrent corneal erosion than controls (0.78 vs 0.66, respectively; p < 0.05). There was a statistically significant difference in the proportion of carriers of the IL10 -592A allele between patients with recurrent corneal erosion and population sample (0.483 vs 0.327, respectively, p < 0.05). Conclusion: IL6 174G/С, IL10 -592С/A and IL10 -1082G/C and the genes determining the pathological processes in the cornea produce a cumulative effect towards modifying the clinical phenotype in keratoconus and lattice corneal dystrophy. Keywords: keratoconus, lattice corneal dystrophy, recurrent erosions, clinical phenotype, interleukin gene polymorphism, genetic predisposition
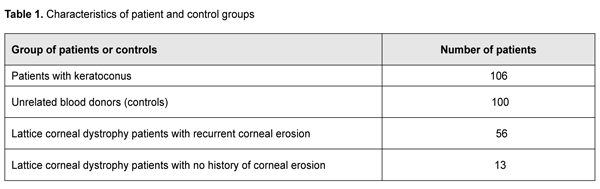
Introduction The disorders associated with pathological changes in the cornea are a challenge for the ophthalmologist, because they substantially affect visual acuity and may lead to visual disability. Keratoconus (or corneal ectasia) is one of the most common corneal disorders with an average annual incidence rate of 2.0 per 100,000 population [1], and prevalence ranging from 50 to 230 cases per 100,00 population, with a wide variation in prevalence across ethnic populations. Keratoconus is recognized as a multifactorial disease with a genetic component, and a number of candidate genes have been linked to the disease. Lattice corneal stromal dystrophy (LCD), a monogenic disorder with varying phenotypic manifestations, is another important issue and the most common hereditary corneal dystrophy in Ukraine, with as much as about 40.2% of hereditary corneal dystrophy cases attributed to this disease. Keratoconus (KC) is an idiopathic, progressive, non-inflammatory corneal dystrophy that is commonly chronic. It is a condition in which the cornea assumes a conical shape because of thinning and protrusion. This results in longitudinal increases in corneal curvature and changes in corneal optical characteristics, leading to incorrect central and paracentral astigmatism with significantly decreased vision. There is accumulating evidence supporting a genetic basis of keratoconus, while an environmental factor may be essential to act as a trigger in genetically predisposed individuals. The bilaterality of the disease strongly supports a genetic basis. In addition, the disease is frequently associated with hereditary disorders or syndromes (Down’s syndrome, Marfan's syndrome, Leber’s blindness, optic nerve atrophy, retinal pigment degeneration, cataract, Crouzon's syndrome, blue sclera, incomplete osteogenesis, type IV Ehlers-Danlos' syndrome, mitral valve prolapse, etc) [2-5]. In addition, a genetic predisposition to keratoconus is well documented with increased incidence in some familial groups [6, 7] and numerous reports of concordance between monozygotic twins [8-11]. It is noteworthy that positive family history has been documented in 6% to 23% KC subjects [12, 13]. First degree family members are at higher risk of keratoconus than normal population [14]. A number of candidate genes have been linked to KC. There is a number of heritable and frequently associated with each other quantitative traits among biometric parameters of the human eye. One of them is central corneal thickness (CСT), with its heritability being as high as 0.95 [15]. Extreme CCT measurements have been associated with rare hereditary connective tissue disorders such as brittle cornea syndrome (BCS) and several types of osteogenesis imperfect [16,17], whereas moderate CCT measurements have been associated with more common eye disorders in elderly individuals. CСT is believed to be an essential trait in keratoconus [18,19]. Previous genome-wide association studies (GWAS) have identified eleven loci associated with ССT in European and Asian populations [20,21,22,23]. Association of KC with certain gene polymorphisms, such as those of hepatocyte growth factor (HGF), has been demonstrated [24]. Although KC is defined as a non-inflammatory corneal dystrophy, recent studies have shown that cytokines, proteolytic enzymes and free radicals play a role in the development of the disease [25]. Elevated tear IL6, TNF-? and ММР-9 levels have been demonstrated in patients with KC. In addition, eye rubbing contributes to increased tear IL6, TNF-? and ММР-9 levels, and is a risk factor for KC [26-31]. Interleukin-10 (IL10) is an anti-inflammatory cytokine, and the IL10 gene is highly expressed in injured corneal epithelium. Mouse models have demonstrated that IL10 -1082 G/A (rs1800896) substantially effects the level and functional activity of the encoded protein. The recognition site for the transcription factor C/EBP disappears in the presence of the A allele in IL10 rs1800896. Findings of experimental studies have confirmed that such a substitution will be associated with reduced expression of the gene and production of the relevant protein [32, 33]. Given that anti-inflammatory cytokines play a crucial role in eliminating inflammatory cells and protecting against corneal ulcers and neovascularization, we have chosen IL10 rs1800896 as a candidate gene for a genetic predisposition to keratoconus. There have been numerous studies on the effect of cytokines on corneal epithelial repair after mechanical or chemical injury [34,35,36]. The corneal epithelial wound healing response is brought about by a complex cascade of events involving cytokine-mediated interactions between the epithelial cells, keratocytes of the corneal stroma, corneal nerves, lacrimal glands, and cells of the immune system [34]. Thus, pro-inflammatory cytokines IL1, IL6 and IL8 regulate proliferation and migration of corneal cells and degradation of collagen residue, and protect the site of injury from bacterial infection [34,37]. Two allelic variants in the IL6 promoter, -597G/A and -174 G/C, have been considered as risk factors for these disorders. These promoter polymorphisms regulating IL6 gene expression are associated with production of the relevant protein and circulating levels of C-reactive protein [38]. Based on the results of the analysis of a regulatory sequence associated with IL6 gene, we chose a single-nucleotide guanine-to-cytosine substitution at position ?174 of the IL6 gene promoter as a subject for further analysis. Based on the results of analysis of the predicted effect of this polymorphism on the structure of transcription factor recognition sites, we found that the single-nucleotide substitution is located in the recognition site of NRD and may result in disappearance of the cis- regulatory site in the promoter region. In an in vitro study [39], the -174C construct showed lower expression than the -174G construct. In addition, it has been demonstrated that the presence of the above single-nucleotide substitution also effects the capacity of the estrogen receptor-estrogen complex to regulate the activity of IL6 gene promoter [40]. We have previously found that cytokine genes are expressed in the corneal epithelium, and their polymorphisms are associated with the risk of and/or protection against recurrent corneal erosion in patients with LCD [41]. Given the importance of IL6 -174 G/C and IL10 -1082G/A and -592C/A polymorphisms for visual function, we have chosen these genetic polymorphisms as candidate SNPs for keratoconus and candidate genes modifying LCD phenotypes. Therefore, the purpose of this study was to elucidate the role of polymorphic variants in the IL6 promoter (-174 G/C) and IL10 (-1082G/A and -592C/A) as factors of a genetic predisposition to keratoconus and recurrent corneal erosion in Ukrainian patients with LCD. Material and Methods Analysis of clinical data and sampling A DNA bank of patients with keratoconus and those with LCD type I or type I/III has been established. The material for the study consisted of blood samples obtained from (a) patients with the above diseases at the Filatov Institute, and (b) blood donors from various Ukrainian regions who were not related to each other and represented the general population of Ukraine. All participants provided informed consent before entering the study. All DNA samples were coded and tested in an anonymous manner. Patients with keratoconus who were treated at the Filatov Institute were included in the study. Results of the control population sample were provided by the Department of Human Genetics, Institute of Molecular Biology and Genetics of the NAS of Ukraine. Table 1.1 presents characteristics of patient groups.
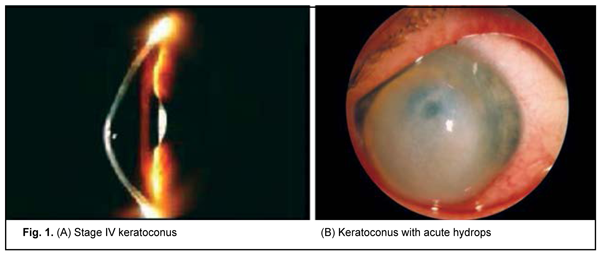
The results of the genetic study in 106 patients with keratoconus (212 eyes; age, 10 years to 67 years; mean age, 31.5±11.82 years) were subjected to analysis. Patients were mostly males (65%). Sixty-nine patients with LCD (46 patients with TGFBI Arg124Cys LCD type I and 23 patients with Hys626Arg LCD type I/IIIA) underwent molecular genetic studies. Of these, 56 had recurrent corneal erosion (RCE) and 13 had no history of corneal erosion. Clinical examination All patients underwent a routine eye examination including uncorrected (UCVA) and best-corrected visual acuity (BCVA) assessment, biomicroscopy, fluorescein testing, tonometry and ophthalmoscopy. In addition, patients with keratoconus underwent keratotopography, pachymetry, remote biometry and gonioscopy. Twenty-four eyes had stage I keratoconus (astigmatism ? 5.0 D; keratometry, ? 48.0 D; visual acuity, 0.5 to 1.0), 52 eyes had stage II (astigmatism, 5.0 to 8.0 D; keratometry, ? 53.0 D; pachymetry, ? 400 ?m; visual acuity, 0.1 to 0.4), 44 eyes had stage III (astigmatism, 8.0 to 10.0 D; keratometry, > 53.0 D; pachymetry, 300 to 400 ?m; visual acuity, 0.12), and 83 eyes had stage IV (keratometry, > 55.0 D; pachymetry, <200 ?m; visual acuity, 0.01 to 0.02) (Amsler-Krumeich classification). Genetic examination Genomic DNA samples were isolated from peripheral blood lymphocytes using proteinase K digestion of the cell lysate and subsequent phenol extraction. Spectral analysis and 0.6% agarose gel electrophoresis were used to determine sample quality. The concentration and spectral characteristics of the DNA sample were determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE USA). PCR was performed automatically on the 2720 thermal cycler (Applied Biosystems, Waltham, MA) or iCycler (Bio-Rad, Hercules, CA). PCR amplification with subsequent hydrolysis of restriction enzyme, NlaIII, and visualization with 2% agarose gel electrophoresis were used for the analysis of the DNA sequence containing the substitution -174 G-->C in the IL6 promoter. The -1082G/A polymorphism in the promoter region of the IL10 gene represents a single-nucleotide G-to-A substitution leading to the disappearance of the recognition site for restriction enzyme EcoNI. Therefore, in vitro PCR amplification with subsequent restriction digestion of PCR products with restriction enzyme, EcoNI, were used for detection of the SNP. After digestion of PCR products with restriction enzyme, EcoNI, they were separated and analyzed by 2% agarose gel electrophoresis. Molecular genetic studies of the IL10 -592 C/A polymorphism were conducted on the basis of PCR and subsequent restriction fragment length polymorphism (RFLP) analysis. The -592 C/A polymorphism represents a single-nucleotide C-to-A substitution in the promoter region of the IL10 gene. As a result of this substitution, the restriction site appears which leads to differential digestion of various allelic variants in the IL10 gene with restriction enzyme, RsaI, making them identifiable. After digestion of PCR amplification products with restriction enzyme, RsaI, they were separated and analyzed by 2% agarose gel electrophoresis. Statistical analyses were conducted using OpenEpi (Andrew G. Dean and Kevin M. Sullivan, Atlanta, GA) software. Chi-square test and Fexact test were used. Results and Discussion Clinical manifestations of keratoconus KC is a progressive and commonly bilateral corneal dystrophy that leads to corneal thinning and deformation and uncorrectable decreased visual acuity. Disease onset commonly occurs in young individuals of working age, and the rate of progression varies between individuals. In patients of this study, signs of disease were first noted at a median age of 19.6 + SD7.01 years (range 6 to 40 years). It is important to differentiate between subclinical KC and clinical KC. Subclinical keratoconus is characterized by the appearance of indirect signs (symptoms) such as complaints of astenopia, formation of myopic (sometimes mixed) astigmatism in pubertal age or beyond it, decreased visual acuity that improves under diaphragming conditions, decreased best-corrected visual acuity, and instability of ophthalmometric measurements on the eyes. Enhanced appearance of corneal nerves and sparse stroma in the paracentral cornea are a biomicroscopic sign of early keratoconus [2]. Clinical disease is characterized by a narrow optic section in the ectactic zone and Vogt's striae, fine vertical stromal lines that are produced by Descemet’s membrane compression. These lines disappear when external pressure is applied to the globe. Other signs appear as the disease progresses, including decreased visual acuity, astigmatism that is difficult or impossible to correct, myopic refractive error combined with astigmatism which is usually incorrect, increased corneal refractive power, small radius of corneal curvature, and corneal thinning. As the cone further grows, the apex becomes scarred and opaque, with thinning involving more and more peripheral cornea [2, 3]. Of the patients enrolled in the current study, most (60%; 127 eyes) had stage III or IV keratoconus. Acute hydrops were seen in 6 eyes (2.8%) (Fig. 1).
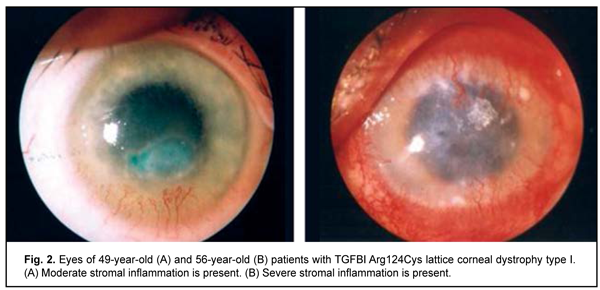
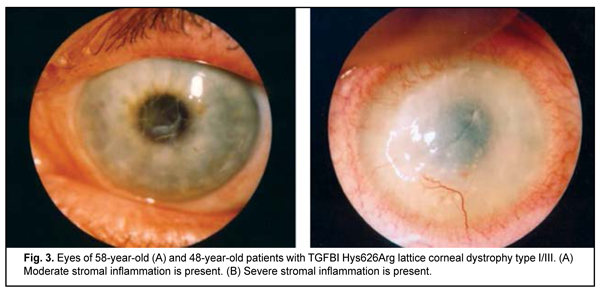
Clinical manifestations of lattice corneal dystrophy The complete clinical picture of lattice dystrophy type 1 generally develops in the third to fourth decade of life. The typical diagnostic triad of LCD comprises (1) transparent double-contoured radially branching lattice structures in the anterior and intermediate stroma; (2) punctate grayish opacities in the superficial stroma; and (3) diffuse, central subepithelial opacity [45, 46]. Radially oriented lattice lines with dichotomous branching near their central terminations are pathognomonic for LCD. Recurrent corneal erosions may be associated with LCD, develop as early as childhood, and are the most common major complaint in patients with LCD attending an eye department. Patients, however, vary in the number of recurrent episodes, intensity of recurrence and severity of the concurrent inflammatory response (Fig. 2 a, b). Epithelial injury is a mechanism triggering the inflammatory response in LCD. The mean age of our patients with LCD type 1 was 41.7 ±SD11.2 years.
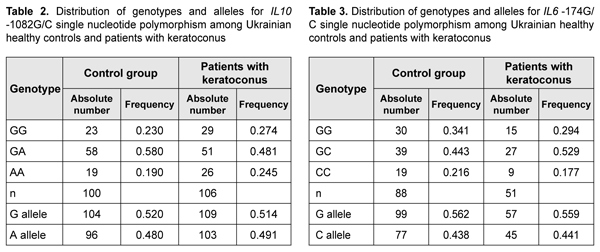
The complete clinical picture of LCD type I/IIIA generally develops in the fourth to fifth decade of life, but the components of the typical diagnostic triad are somewhat less apparent than in LCD type I. LCD type I/IIIA is characterized by thinner lattice structures compared to LCD type I, and asymmetric manifestations with no or rare recurrence of corneal erosion [46]. The mean age of our patients with LCD type I/IIIA was 55.3±SD9.6 years, which was older that the mean age of patients with LCD type I due to later manifestation of this LCD type. Results of studies of IL6 and IL10 polymorphisms in patients with keratoconus and LCD IL10 rs1800896 was analyzed in 106 patients and 100 healthy controls. Table 2 presents the distribution of genotypes and alleles for IL10 rs1800896.
Patients with keratoconus exhibited higher frequency of the homozygous (AA) genotype of rs1800896 than controls (25% vs 19%, respectively). Although the difference was not significant, but there was a tendency for association between the homozygous (AA) genotype of IL10 rs1800896 and keratoconus. Frequency of the homozygous (CC) genotype of IL6 -174 G/C was higher in control subjects (22%) compared to patients with keratoconus (18%). Although the difference was not significant, but there was a tendency for association between the homozygous (CC) genotype of IL6 -174 G/C and keratoconus. A recent meta-analysis of genome-wide association studies identified 11 keratoconus-associated loci [42]. The findings we obtained may be considered as potential evidence of an additive cumulative effect of our investigated SNPs of IL6 and IL10, and the candidate genes that have been identified by the meta-analysis. To study such a cumulative modifying effect of the IL6 and IL10 SNPs on the clinical phenotype in patients with another corneal disorder, particularly, lattice corneal dystrophy, we investigated polymorphisms of these genes in patients with this diagnosis. Hereditary stromal corneal dystrophies (particularly, lattice stromal corneal dystrophy) are a group of autosomal-dominant disorders caused by mutations in the transforming growth factor, beta-induced (TGFBI) gene [43,44]. We conducted molecular genetic studies of these variants in 56 patients with recurrent corneal erosions and 13 patients with no history of corneal erosion. Previously, we have identified the TGFBI gene mutations characteristic for lattice dystrophy in these patients [45].
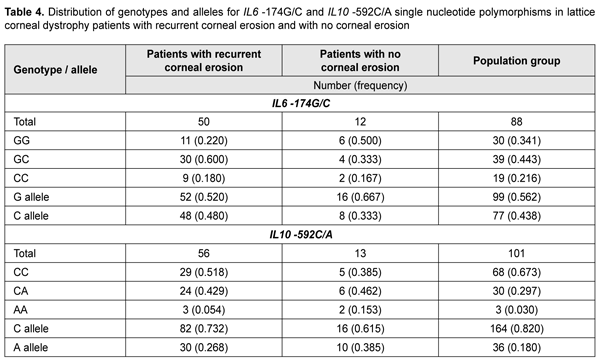
To elucidate the possible role of IL6 -174G/С and IL10 -592С/A (the latter SNP is non-equilibrium linked with IL10 -1082G/C that we have already investigated) as phenotype modifiers in patients with LCD, we conducted molecular genetic studies of these variants in patients with recurrent corneal erosions. Table 4 presents the distribution of genotypes among groups.
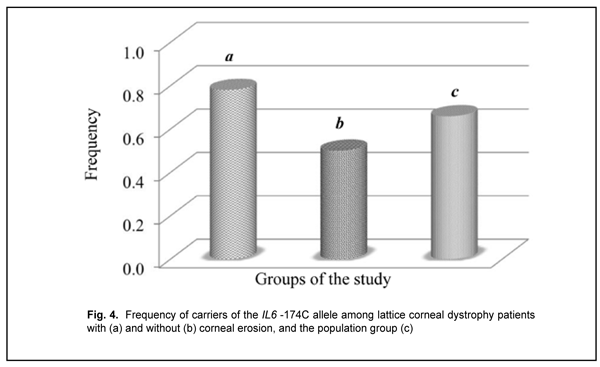
Among the group of patients with corneal erosions, we found 9 individuals (0.18) who were homozygous for the –174C allele that had been shown to be associated with a decreased production of IL6. The heterozygous -174GC genotype was a major genotype (0.600) both in the group and the population sample under study. A deviation from Hardy–Weinberg equilibrium, evaluated by comparing observed and expected genotype frequencies with an exact goodness of fit test, was observed in the patients with recurrent corneal erosion associated with LCD for this polymorphic variant (?2=4.08; р=0.04). Distribution of allele frequencies for the polymorphic variant was characterized based on the distribution of genotype frequencies among study patients and two control groups. The frequency of the carriers of the IL6 -174C allele was 0.780 in the corneal erosion group compared to 0.659 in the control population sample (p < 0.05) (Fig. 4) and 0.500 in the no-erosion control group; the difference for the latter group, however, did not reach statistical significance (p > 0.05), likely due to small group size.
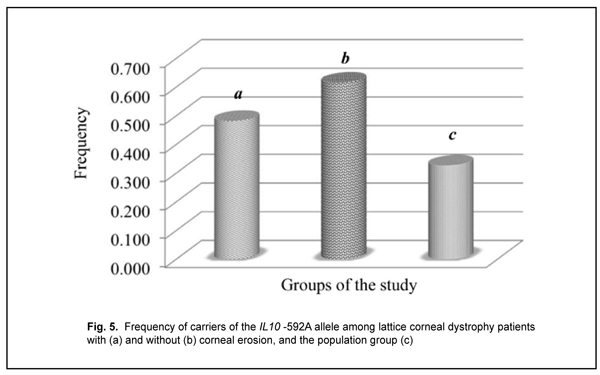
The distribution of genotypes for the polymorphic allele of IL10 -592 С/A among the study group and control group was determined based on the results of molecular genetic studies. The homozygous wild-type (-592 СС) genotype was the most common (0.518) for patients with recurrent corneal erosion and population sample, whereas the heterozygous (-592 СA) genotype was the most common (0.462) for patients without corneal erosions. No deviation from Hardy–Weinberg equilibrium, evaluated by comparing observed and expected genotype frequencies with an exact goodness of fit test, was observed in the patients with recurrent corneal erosion associated with LCD for this polymorphic variant (?2=0.49; р=0.380). There was a statistically significant difference (p < 0.05) in the proportion of carriers of the IL10 -592A allele between patients with recurrent corneal erosion (0.483) and population sample (0.327) (Fig. 5).
This may be explained given that recurrent corneal erosions may differ in the degree of concomitant inflammation [34]. Impaired adhesion of basal epithelial cells to the Bowman layer due to accumulation of amyloid deposits (abnormal protein fibers) in between these structures, and secondary degenerative changes in the anterior stroma provide the morphological basis for recurrent corneal erosions in LCD [46]. Epithelial injury is a mechanism triggering the inflammatory response in LCD. Corneal regeneration is a complex body response, with its success and timing depending on the correct combination of cytokines and growth factors expressed at specific times [34,47,35]. Pro-inflammatory cytokines co-regulate proliferation of corneal cells and degradation of necrotized cells and denaturated collagen [36,48]. Anti-inflammatory cytokines provide elimination of inflammatory cells, thus preventing corneal ulceration, melting and neovascularization [34,47]. The genes of the two major cytokines of acute inflammation (IL1? and IL6), chemokine IL8 and anti-inflammatory cytokine IL10 are expressed in the injured corneal epithelium [36,49]. The proteins encoded by these genes have been found in the tear fluid of patients with injured corneal tissue and shown to play a key role in corneal regeneration [37]. It may be assumed that, in patients with lattice corneal dystrophy, the degree of inflammation that rises in recurrent erosions is under control of modifying genes. Conclusion We analyzed, for the first time, the distribution of genotypes and alleles for IL6 -174G/С and IL10 -592С/A in patients with keratoconus and those with recurrent erosions (including those with and without lattice corneal dystrophy). We found that IL6 -174С and IL10 -592A alleles are potential inflammation modifiers and potential genetic risk factors for recurrent corneal erosions in patients with lattice corneal stromal dystrophy. The frequency of homozygotes (AA) for IL10 rs1800896 was increased, whereas the frequency of homozygotes (CC) for IL6 174G/С was decreased in patients with keratoconus compared to controls (0.25 vs 0.19 and 0.18 vs 0.22, respectively), although the differences were not statistically significant. Given the multifactorial pathogenesis of keratoconus, further research with substantially increased sample sizes is warranted to achieve statistically significant differences and verify these findings. Therefore, we obtained one more confirmation of an additive cumulative effect of IL6 174G/С, IL10 -592С/A and IL10 -1082G/C and the genes determining the pathological processes in the cornea.
References 1.Kennedy RH, Bourne WM, Dyer JA. A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986 Mar 15;101(3):267-73. 2.Dronov MM, Pirogov IuI. [Keratoconus: Types, Etiology and Pathogenesis. Part 1]. Oftalmokhirurgiia i terapiia. 2002;2:33-7. Russian. 3.Sevost'ianov EN, Gorskova EN, Ekgardt VF. [Keratoconus (ethiology, pathogenesis, and medicinal treatment): a tutorial]. Cheliabinsk: UGMADO; 2005. Russian. 4.Birich TA, Chekina AIu, Aksenova NI. [Treatment outcomes for patients with keratoconus]. Oftalmologiia Belarusi. 2010;1(4). Russian. 5.Nowak DM, Gajecka M. The genetics of keratoconus. Middle East Afr J Ophthalmol. 2011 Jan;18(1):2-6. 6.Karimian F, Aramesh S, Rabei HM, Javadi MA, Rafati N. Topographic evaluation of relatives of patients with keratoconus. Cornea. 2008 Sep;27(8):874-8. 7.Owens H, Gamble G. A profile of keratoconus in New Zealand. Cornea. 2003;22:122–25. 8.Aknin C, Allart JF, Rouland JF. [Unilateral keratoconus and mirror image in a pair of monozygotic twins]. J Fr Ophtalmol. 2007 Nov;30(9):899-902. 9.Weed KH, MacEwen CJ, McGhee CN. The variable expression of keratoconus within monozygotic twins: dundee University Scottish Keratoconus Study (DUSKS). Cont Lens Anterior Eye. 2006 Jul;29(3):123-6. 10.Schmitt-Bernard C, Schneider CD, Blanc D, Arnaud B. Keratographic analysis of a family with keratoconus in identical twins. J Cataract Refract Surg. 2000 Dec;26(12):1830-2. 11.Parker J, Ko WW, Pavlopoulos G, Wolfe PJ, Rabinowitz YS, et al. Videokeratography of keratoconus in monozygotic twins. J Refract Surg. 1996 Jan-Feb;12(1):180-3. 12.Hughes AE, Dash DP, Jackson AJ, Frazer DG, Silvestri G. Familial keratoconus with cataract: linkage to the long arm of chromosome 15 and exclusion of candidate genes. Invest Ophthalmol Vis Sci. 2003 Dec;44(12):5063-6. 13.Rabinowitz YS. The genetics of keratoconus. Ophthalmol Clin North Am. 2003 Dec;16(4):607-20, vii. 14.Wheeler J, Hauser MA, Afshari NA, Allingham RR, Liu Y. The Genetics of Keratoconus: A Review. Reprod Syst Sex Disord. 2012 Jun 3;(Suppl 6). 15.Dimasi DP, Burdon KP, Craig JE. The genetics of central corneal thickness. Br J Ophthalmol. 2010 Aug;94(8):971-6. 16.Pedersen U, Bramsen T. Central corneal thickness in osteogenesis imperfecta and otosclerosis. ORL J Otorhinolaryngol Relat Spec. 1984;46(1):38-41. 17.Evereklioglu C, Madenci E, Bayazit YA, et al. Central corneal thickness is lower in osteogenesis imperfecta and negatively correlates with the presence of blue sclera. Ophthalmic Physiol Opt. 2002 Nov;22(6):511-5. 18.Cohen EJ, Myers JS. Keratoconus and normal-tension glaucoma: a study of the possible association with abnormal biomechanical properties as measured by corneal hysteresis. Cornea. 2010 Sep;29(9):955-70. 19.Gordon M.O, et al. The Ocular Hypertension Treatment Study: baseline factors that predict the onset of primary open-angle glaucoma. Arch Ophthalmol. 2002 Jun;120(6):714-20; discussion 829-30. 20.Cornes BK, Khor CC, Nongpiur MI, et al. Identification of four novel variants that influence central corneal thickness in multi-ethnic Asian populations. Hum Mol Genet. 2012 Jan 15;21(2):437-45. 21.Lu Y, Dimasi DP, Hysi PG, et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010 May 13;6(5):e1000947. 22.Vitart V, Benci? G, Hayward C, et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum Mol Genet. 2010 Nov 1;19(21):4304-11. 23.Vithana EN, Aung T, Khor CC, et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 2011 Feb 15;20(4):649-58. 24.Burdon KP, Macgregor S, Bykhovskaya Y, et al. Association of Polymorphisms in the Hepatocyte Growth Factor Gene Promoter with Keratoconus. Invest Ophthalmol Vis Sci. 2011 Oct 31;52(11):8514-9. 25.Chwa M, Atilano SR, Reddy V, Jordan N, Kim DW, Kenney MC. Increased stress-induced generation of reactive oxygen species and apoptosis in human keratoconus fibroblasts. Invest Ophthalmol Vis Sci. 2006 May;47(5):1902-10. 26.Balasubramanian SA, Mohan S, Pye DC, Willcox MD. Proteases, proteolysis and inflammatory molecules in the tears of people with keratoconus. Acta Ophthalmol. 2012 Jun;90(4):e303-9. 27.Lema I, Duran JA. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology. 2005 Apr;112(4):654-9. 28.Lema I, Sobrino T, Duran JA, Brea D, Diez-Feijoo E. Subclinical keratoconus and inflammatory molecules from tears. Br J Ophthalmol. 2009 Jun;93(6):820-4. 29.Nemet AY, Vinker S, Bahar I, Kaiserman I. The association of keratoconus with immune disorders. Cornea. 2010 Nov;29(11):1261-4. 30.Wisse RP, Kuiper JJ, Gans R, Imhof S, Radstake TR, Van der Lelij A. Cytokine expression in keratoconus and its corneal microenvironment: a systematic review. Ocular Surf. 2015 Oct;13(4):272-83. 31.Jun AS, Cope L, Speck C, et al. Subnormal cytokine profile in the tear fluid of keratoconus patients. PLoS One. 2011 Jan 27;6(1):e16437. 32.Temple S, Lim E, Cheong K, et al. Alleles carried at positions ?819 and ?592 of the IL10 promoter affect transcription following stimulation of peripheral blood cells with Streptococcus pneumoniae. Immunogenetics. 2003;55:629–32. 33.Costa GC, da Costa Rocha MO, Moreira PR, et al. Functional IL-10 Gene Polymorphism Is Associated with Chagas Disease Cardiomyopathy. J Infect Dis. 2009 Feb 1;199(3):451-4. 34.Agrawal VB, Tsai RJ. Corneal epithelial wound healing. Indian J Ophthalmol. 2003 Mar;51(1):5-15. 35.Wilson SE, Mohan RR, Ambr?sio R, et al. The corneal wound healing response: cytokine-mediated interaction of the epithelium, stroma, and inflammatory cells. Prog Retin Eye Res. 2001 Sep;20(5):625-37. 36.Ebihara N, Matsuda A, Nakamura S, et al. Role of the IL-6 classic- and trans-signaling pathways in corneal sterile inflammation and wound healing. Invest Ophthalmol Vis Sci. 2011 Nov 1;52(12):8549-57. 37.Sotozono C, He J, Matsumoto Y, et al. Cytokine expression in the alkali-burned cornea. Curr Eye Res. 1997 Jul;16(7):670-6. 38.Ferrari SL, Ahn-Luong L, Garnero P, et al. Two promoter polymorphisms regulating interleukin-6 gene expression are associated with circulating levels of C-reactive protein and markers of bone resorption in postmenopausal women. J Clin Endocrinol Metab. 2003 Jan;88(1):255-9. 39.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998 Oct 1;102(7):1369-76. 40.Kristiansen OP, Nols?e RL, Larsen L, et al. Association of a functional 17beta-estradiol sensitive IL6-174G/C promoter polymorphism with early-onset type 1 diabetes in females. Hum Mol Genet. 2003 May 15;12(10):1101-10. 41.Kucherenko AM, Pampukha VM, Drozhzhyna GI, Livshits LA. IL1beta, IL6 and IL8 gene polymorphisms involvement in recurrent corneal erosion in patients with hereditary stromal corneal dystrophies. Tsitol Genet. 2013 May-Jun;47(3):42-5. 42.Yi Lu, Vitart V, Burdon KP, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013 Feb;45(2):155-63. 43.Aldave A. The genetics of the corneal dystrophies. Dev Ophthalmol. 2011. 2011;48:51-66. 44.Kannabiran C, Klintworth GK. TGFBI gene mutations in corneal dystrophies. Hum Mutat. 2006 Jul;27(7):615-25. 45.Pampukha VM, Drozhyna GI, Livshits LA. TGFBI gene mutation analysis in families with hereditary corneal dystrophies from Ukraine. Ophthalmologica. 2004 Nov-Dec;218(6):411-4. 46.Weidle E. Epithelial and stromal corneal dystrophies. Ophthalmologe. 1996; 93(6):754–67. 47.Reinach P, Pokorny PS. The corneal epithelium: clinical relevance of cytokine-mediated responses to maintenance of corneal health. Arq Bras Oftalmol. 2008 Nov-Dec;71(6 Suppl):80-6. 48.Hong JW, Liu JJ, Lee JS, et al. Proinflammatory chemokine induction in keratocytes and inflammatory cell infiltration into the cornea. Invest Ophthalmol Vis Sci. 2001 Nov;42(12):2795-803. 49.Torres P, Kijlstra A. The role of cytokines in corneal immunopathology. Ocul Immunol Inflamm. 2001 Mar;9(1):9-24.
The authors certify that they have no conflicts of interest in the subject matter or materials discussed in this manuscript.
|