J.ophthalmol.(Ukraine).2017;2:3-7.
|
https://doi.org/10.31288/oftalmolzh2017237 Relationship of the AKR1B1 rs759853 and rs9640883 with the development of diabetic retinopathy
S.Iu. Mogilevskyy1, Dr Sc (Med), Prof. O. V. Bushuieva2, Assistant S.V. Ziablitsev3, Dr Sc (Med), Prof. L.V. Natrus3, Dr Sc (Med), Prof. 1Shupik National Medical Academy of Postgraduate Education 2Danylo Halytsky Lviv National Medical University 3Bohomolets National Medical University Kyiv, Lviv, Ukraine E-mail: sergey.mogilevskyy@gmail.com
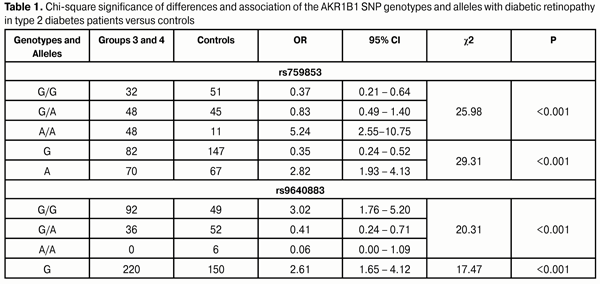
Background: In type 2 diabetes mellitus (DM2), hyperglycemia activates the polyol pathway of glucose metabolism, resulting in accumulation of sorbitol and fructose. Polymorphism of aldose reductase (a key enzyme of the poliol pathway) gene (AKR1B1) influences the activity of the polyol pathway and might be associated with the development of diabetic retinopathy (DR). Purpose: To identify any association of the polymorphisms influencing the activity of aldose reductase gene (rs759853 and rs9640883 of AKR1B1) with DR in the Ukrainian population. Materials and Methods: In total, 311 individuals were involved in the study. Group 1 (control group) was composed of 107 ophthalmologically normal subjects. Group 2 contained 76 patients with stage 1 DR (i.e., showing no fundus changes). Group 3 consisted of 64 patients diagnosed with non-proliferative DR (NPDR), and Group 4 was composed of 64 patients diagnosed with proliferative DR (PDR). The TaqMan Mutation Detection Assays were used to analyze polymorphic DNA loci in a Real-Time PCR System 7500. Results and Discussion: In Ukrainian population, rs759853 and rs9640883 of AKR1B1 were associated with the development of DR. The mutant A allele and AA genotype at the rs759853 increased the odds (OR=2.8 and OR=5.2, respectively; р<0.001) of developing DR. In addition, the ancestral G allele and the homozygous G/G status at the rs9640883 increased the odds (OR=2.6 and OR=3.0, respectively; р<0.001) of developing DR. The presence of risk alleles had a negative effect on the development of DR in DM2. The AKR1B1 rs759853 and rs9640883 had no effect on the development of PDR. Importance of the polymorphisms for the development of DR was demonstrated by dispersion analysis (rs759853: F = 18.9, p < 0.001; rs9640883: F = 6.9, p = 0.001). Conclusion: The frequency of the ancestral G allele of the AKR1B1 rs759853 significantly decreased, whereas that of the mutant A allele increased with increased severity of pathological process in DR. The frequency of the ancestral GG genotype of the AKR1B1 rs9640883 was significantly elevated, whereas that of the heterozygous GA genotype was decreased, and minor homozygous AA genotype was absent in diabetes patients with DR. Key words: diabetic retinopathy, AKR1B1, rs759853, rs9640883 Introduction Genetic factors are no doubt important in the development of diabetic retinopathy (DR), and are thought to account for up to 50% of the risk for the disorder [1-4]. Identification of patients predisposed to develop DR would facilitate the development of individualized approaches to prevention and treatment. In addition, genetic factors can be considered as essential for the development of various complications of type 2 diabetes mellitus (DM2), including, first and foremost, DR [5, 6]. It has been demonstrated in different populations that heredity has a role in the development of DR, and that this role does not depend on the hyperglycemia level and on other environmental factors contributing to the risk [7, 8]. The mechanism of the development of DR involves numerous components, including alterations in hydrocarbon, lipid, protein and electrolyte metabolism. Hyperglycemia activates the polyol pathway of glucose metabolism, resulting in accumulation of sorbitol and fructose. In the absence of hyperglycemia, the percentage of glucose converted to sorbitol is not more than 1%; however, the percentage increases to 7-8% in DM2 patients [5]. Aldose reductase, a key enzyme of the poliol pathway, reduces glucose in sorbitol. It is the very enzyme in which dysregulation of intracellular homeostasis begins, as sorbitol and fructose, the products of the sorbitol pathway of glucose metabolism, do not diffuse readily through cell membranes and accumulate intracellularly [8]. Therefore, investigating the association of the polymorphisms influencing the activity of aldose reductase gene (rs759853 and rs9640883 of AKR1B1) with DR in the Ukrainian population is very important. The study purpose was to identify any association of the AKR1B1 rs759853 and rs9640883 with the DR in the Ukrainian population. Materials and Methods The study was conducted at the Ophthalmology Department of the Danylo Halytsky Lviv National Medical University. Group 1 (control group) was composed of 107 ophthalmologically normal subjects (age- and sex-matched with patients) having neither diabetes mellitus nor any other systemic disease. Group 2 contained 76 patients with DM2 showing no fundus changes. Group 3 consisted of 64 patients diagnosed with non-proliferative DR (NPDR), and Group 4 was composed of 64 patients diagnosed with proliferative DR (PDR). All patients of groups 2, 3 and 4 had undergone cataract surgery. In total, 311 individuals were involved in the study. Ophthalmological examination included visual acuity assessment, Goldmann tonometry, static Humphrey perimetry (Carl Zeiss Meditec, Dublin, CA), slit lamp biomicroscopy (BQ 900; Haag-Streit, K?niz, Switzerland); gonioscopy; ophthalmoscopy with contact and non-contact lenses (Volk Optical, Mentor, OH); optical coherence tomography (Optovue, Inc., Fremont, CA); and fundus photography (the ETDRS seven standard fields) and fluoresecent angiography with the fundus camera TRC-NW7SF (Topcon, Tokyo, Japan). TaqMan Mutation Detection Assays (Thermo Fisher Scientific) were used to analyze polymorphic DNA loci in a Real-Time PCR System (7500 system; Applied Biosystems, Foster City, CA). The rs759853 polymorphism of the AKR1B1 gene is located on chromosome 7q35 (Chr. 7:134143958 as per NCBI Build 37). This is a G-to-A SNP in an intron of the AKR1B1 (NM_001628.2: c.-144 C>T). Here G is an ancestral allele, A is a minor allele with minor allele frequency = 0.2768 according to MAF Source (1000 Genomes (http://www.1000genomes.org/node/506).) The rs9640883 polymorphism of the AKR1B1 gene is located in Chr. Chr.7:134116633 as per NCBI Build 37. This is a G-to-A SNP in an intron of the AKR1B1 (XR_928003.1: n.94+433 C>T). Here G is an ancestral allele, A is a minor allele with minor allele frequency = 0.2853 (MAF Source: 1000 Genomes http://www.1000genomes.org/node/506). Statistical analysis of the results of clinical studies was performed using SPSS 11.0 Statistical Software Package (SPSS, Inc, Chicago, IL), Medstat Software (Iu.Ie. Liakh. G.V. Gur’ianov), and MedCalc (MedCalc SoftWare bvba, 1993-2013). A P value of <0.05 was considered statistically significant in all analyses. Results and Discussion In the first phase of the study, the effect of the SNPs on the development of DR in patients with DM2 was investigated. With this in mind, we investigated whether there was any association between the genotypes and alleles of the SNPs and DR in Groups 3 and 4 versus Group 1 (controls) (Table 1). The table demonstrates that both SNPs were strongly associated with the development of DR in patients with DM2 (p < 0.001 in all cases).
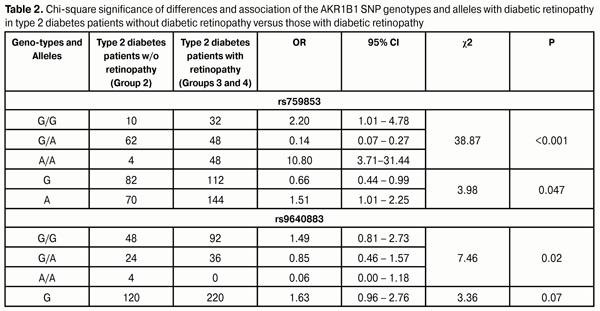
The homozygous GG genotype at rs759853 was associated with a 2.7-fold decreased risk (OR=0.37; 95% CI: 0.21-0.64), whereas the homozygous AA genotype at rs759853 was associated with a 5.2-fold increased risk (OR=5.24; 95% CI: 2.55-10.75) for DR in cases compared to controls. In addition, analysis of the multiplicative inheritance model demonstrated a similar association of DR with allelic polymorphism: the ancestral G allele was associated with a 2.9-fold decreased risk (OR=0.35; 95% CI: 0.24-0.52) whereas the mutant A allele was associated with a 2.8-fold increased risk (OR=2.82; 95% CI: 1.93-4.13) for the disease. The homozygous GG genotype at rs9640883 was at 3.0-fold increased risk (OR = 3.02; 95% CI: 1.76-5.20) for the development of DR. The heterozygous GA genotype or minor homozygous AA genotype of this SNP was associated with a 2.4-fold decreased risk (OR=0.41; 95% CI: 0.24-0.71). Analysis of the multiplicative inheritance model demonstrated that the wild-type G allele of rs9640883 was associated with a 2.9-fold increased risk (OR=2.61; 95% CI: 1.65-4.12), whereas the minor A allele was associated with a 2.6-fold decreased risk (OR=0.38; 95% CI: 0.24-0.61) for the disease. In the next phase of the study, patients with DM2 without DR (Group 2) were compared against patients with DM2 with DR (Group 3 plus Group 4; Table 2). The data obtained demonstrated that the distribution of genotypes of the SNPs under investigation was associated with the presence of DR in patients with DM2. The homozygous rs759853 genotypes, GG and AA, increased the chance of developing DR by 2.2-fold and 10.8-fold, respectively (p < 0.001; Table 2). The presence of the A allele at this SNP increased the odds of developing DR in patients with DM2 1.5-fold (OR=1.51; 95% CI: 1.01-2.25; р=0.047). As would be expected, the distribution of genotypes in rs9640883 was also associated with DR (р = 0.02).
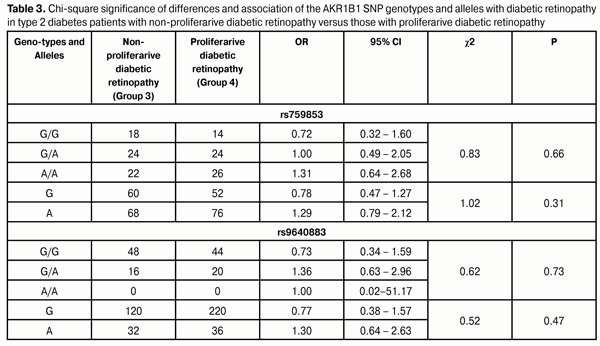
Therefore, the presence of risk alleles in patients with DM2 had a negative effect on the development of DR. The presence of the A allele at rs759853 increased the odds of developing DR in patients with DM2 1.5-fold. Moreover, our study showed a 10.8-fold increased risk of DR in DM2 patients homozygous for the A allele as compared to patients without the allele. These facts suggest that the presence of risk alleles in DM2 patients is a risk factor for incident DR as well as for subsequent increased progression of DR. To identify the role of these SNPs in the incident PDR, we analyzed the difference in genotype and allele distribution among patients with different stages of DR (Table 3). The NPDR and PDR groups were involved into the study to elucidate the effect of polymorphism on the development of proliferative retinal changes in DM2. The data obtained demonstrated that there was no significant difference in distribution of genotypes of the SNPs under investigation among the NPDR and PDR patients. Therefore, one may conclude that the AKR1B1 rs759853 and rs9640883 had no effect on the development of PDR.
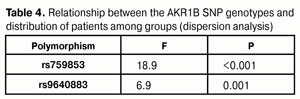
Dispersion analysis was performed to investigate whether there was an association between the polymorphisms and distribution of patients among groups (Table 4). High dispersion coefficients for both AKR1B1 SNPs (rs759853: F = 18.9, p < 0.001; rs9640883: F = 6.9, p = 0.001) indicated the significance of genotype in formation of the groups. Therefore, one may conclude that it was genotype that (a) determined whether a patient had PDR or NPDR, and, in this way, (b) had an effect on the stage of pathological process.
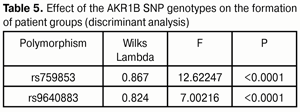
Discriminant analysis (Table 5) demonstrated that rs759853 had a greater effect (F = 12.62) on the distribution of patients among groups compared to rs9640883 (F = 7.00).
Conclusions First, we found that the mutant A allele and AA genotype at the rs759853 increased the odds (OR=2.8 and OR=5.2, respectively; р<0.001), whereas the presence of the G allele in homozygous or heterozygous status significantly decreased the odds of developing DR in DM2, suggestive of a protective effect. In addition, the ancestral G allele and the homozygous G/G status at the rs9640883 increased the odds (OR=2.6 and OR=3.0, respectively; р<0.001), whereas the presence of the A allele and the homozygous AA status at this SNP decreased the odds of developing DR in DM2 by 2.6 times and 16.7 times, respectively (р<0.001). Second, the presence of risk alleles had a negative effect on the development of DR in DM2. The presence of the A allele and the homozygous AA status at the rs759853 increased the odds (OR=1.5 and OR=10.8, respectively) of developing DR in DM2. In addition, the homozygous G/G status at the rs9640883 increased the odds (OR=1.5) of developing DR in DM2. The AKR1B1 rs759853 and rs9640883 had no effect on the development of PDR. Third, high dispersion coefficients for both AKR1B1 SNPs (rs759853: F = 18.9, p < 0.001; rs9640883: F = 6.9, p = 0.001) indicated the significance of genotype in formation of the groups. Finally, discriminant analysis demonstrated that rs759853 had a greater effect (F = 12.62) on the distribution of patients among groups compared to rs9640883 (F = 7.00).
References 1. Abhary S, Hewitt AW, Burdon KP, Craid JE. A systematic meta-analysis of genetic association studies for diabetic retinopathy. Diabetes. 2009; 58(9):2137-47 2. Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007; 445:881-5 3. Hallman DM, Huber JC, Gonzalez VH, et al. Familial aggregation of severity of diabetic retinopathy in Mexican Americans from Starr County, Texas. Diabetes Care. 2005;28(5);1163-8 4. Liew G, Klein R, Wong TY. The role of genetics in susceptibility to diabetic retinopathy. Int Ophthalmol Clin. 2009;49(2):35-52 5. Pan’kiv VI. [Workshop No. 132. Diabetes mellitus: Diagnostic criteria, etiology and pathogenesis]. IEJ. 2013;8:53-64 Ukrainian 6. Cho H, Sobrin L. Genetics of diabetic retinopathy. Curr Diab Rep. 2014;14(8):515. 7. Hietala K, Forsblom C, Summanen P, Groop PH. Heritability of proliferative diabetic retinopathy. Diabetes. 2008;57:2176-80 8. Zhang X, Gao Y, Zhou Z, et al. Familial clustering of diabetic retinopathy in Chongqing, China, type 2 diabetic patients. Eur J Ophthalmol. 2010; 20(5):911-8 9. Suzen S, Buyukbingol E. Recent studies of aldose reductase enzyme inhibition for diabetic complications. Curr Med Chem. 2003;10(15):1329-52
|